This analysis is inspired by Jonathan Dinu's excellent post: https://github.com/hopelessoptimism/happy-healthy-hungry/blob/master/h3.ipynb
Motivation¶
Have you ever seen one of these signs?

Have you ever wondered what it means?
Is this a safe place to eat?
What does a score of 90 mean?
In this lecture we will explore food safety ratings for San Francisco to get a better picture of what these ratings mean.
Thinking about Questions¶
Before we start, let's spend a few minutes thinking about the questions that we want to answer.
What is a good restaurant score for food safety inspections?
What are common issues with restaurants that could be better addressed?
Are there regions of the city sub-par?
Is the situation getting worse or better?
What are some other questions that we might want to answer?
Where can we get data?¶
Description of inspection data for SF:
https://www.sfdph.org/dph/EH/Food/Score/
Data Links from Yelp LIVES program:¶
- Yelp LIVES site: https://www.yelp.com/healthscores
- Data description: https://www.yelp.com/healthscores#businesses
- Downloading data from various locations: https://www.yelp.com/healthscores/feeds
Getting the data¶
I downloaded several feeds and stored them in the local data folder.
Lets first see what files we downloaded data/san_francisco
ls -lh data/san_francisco/
There is data for other cities as well but we will focus on San Francisco for today.
Questions¶
- How big is the data?
- File types?
Looking at the data¶
Data scientists will often use many tools to better understand the data.
The first set of tools we might use are command line utilities:
head -n 5: read the first 5 lines of a filewc: Compute the number of lines (records) in a file.
We can use these command line utilities from within the python notebook. This is done by prefixing each command with the "bang" operator !
Using Head¶
Using head gives us a better picture of the contents of the businesses.csv file:
! head data/san_francisco/businesses.csv
!head -n 3 ./data/san_francisco/inspections.csv
!head -n 3 ./data/san_francisco/violations.csv
Use cat to read the entire file
!cat ./data/san_francisco/legend.csv
The legend indicates what ranges of scores mean. For example, this suggests that poor quality restaurants will have scores at or below 70.
What is Jupyter¶
Environment for describing analysis and doing analysis
Use man to learn¶
To learn more about these commands try:
man cat
# man cat
press esc to clear the pane.
What have we learned so far?¶
- The first line of the file defines the header of the table.
- These files consist of comma separated values (CSV).
- All the fields appear to be quoted (this will be helpful in parsing the file).
- The
business_idfield seems to appear in multiple files and is probably unique in businesses? - We can get a sense for the kind of information in each file (though we should definitely read the documentation)
How big is the data?¶
We already looked at the raw file size:
!ls -Rlh data/san_francisco/
- Why might we want to know the size of the data?
- Why don't we just load it into python?
This dataset is fairly small but in general the data we study may be large (e.g., too big to easily manipulate in memory).
How Many Record?¶
- How many records or observations do we have?
- Can we answer this question just from the size of the file?
In the following we write a small bash script to calculate the number of lines each file.
%%bash
# We can use ipython Magic commands to
# evaluate the entire cell in bash
for f in $(ls -R data/san_francisco/*); do
wc $f
done;
The wc command prints the number of lines, words, and characters respectively in the file.
To learn more about this command run:
man wc
Press esc to close the popup pane.
Counting Lines in Python (Skip in Class)¶
In DS100 we will be using Python to do a lot of our analysis.
How could we write a similar program in Python?
for f in $(ls -R data/san_francisco/*); do
wc $f
done;
import os
for fname in os.listdir("data/san_francisco"):
with open(os.path.join("data/san_francisco", fname), "rb") as f:
lines = sum(1 for line in f)
print("Lines in", fname, ":", lines)
The above Python program demonstrates a few big ideas:
- programmatically traversing the file systems with
os.listdir - using a
withexpression to open (and then automatically close) a file. - using a generator (the expression
sum(1 for line in f)) to count the number of lines without first loading the entire file in memory.
A quick note on file size¶
When we load data into a program it will often increase in size. Therefore we need to be careful when loading entire files:
import sys
with open("data/san_francisco/violations.csv", "r") as f:
lines = f.readlines()
print("Size of list container:",
sys.getsizeof(lines) / (10. ** 6) ,
"MBytes")
print("Sum of lines objects:",
sum(sys.getsizeof(l) for l in lines) / (10. ** 6),
"MBytes")
!ls -Rlh data/san_francisco/violations.csv
First note that the file takes a little more space in memory than it did on disk. Why?
This is because the data structures to store the contents of the file may contain extra fields (e.g., length and points) that add additional overhead.
Data scientists need to think about computational issues:
- Can we fit the data in memory?
- How long will this analysis take to run?
This is why knowing the size of the file can be important before loading it.
Iterating (Skip in Class)¶
What if I wanted to know the number of unique businesses in the violations.csv file without loading the entire file into memory at once?
business_ids = set()
with open("data/san_francisco/violations.csv", "r") as f:
next(f)
for line in f:
business_ids.add(int(line.split(",")[0]))
print("There are", len(business_ids), "unique business ids.")
Interesting there are fewer businesses in the violations.csv file than there are lines in the businesses.csv file. Perhaps there are no violations for some businesses? Let's check to see how many unique business_ids are in the businesses.csv file.
To do this we will practice using bash commands instead.
A little more with bash commands¶
Could we have computed the number of unique businesses using just Bash commands?
tailreturns the end of the file.skips the first line of the file.tail -n +2
catreads the entire filecutextracts a field from row.extracts the first field from a line delimited by commas.cut -f 1 -d ","
sortsorts the inputuniqreturns the unique entries form a sorted inputwcas earlier counts the entries in the output.
We can compose multiple operations using | (pipe).
%%bash
tail -n +2 data/san_francisco/businesses.csv | head -n 5
%%bash
tail -n +2 data/san_francisco/businesses.csv \
| cut -f 1 -d "," \
| head -n 10
The above error occurs because the file is encoded in a non UTF-8 string format. In general this is often the older ISO format ISO-8859-1. We can use iconv to clean up the file.
man iconv
!tail -n +2 data/san_francisco/businesses.csv \
| iconv -t UTF-8 -f ISO-8859-1 \
| cut -f 1 -d "," \
| head -n 10
Adding sort:
%%bash
tail -n +2 data/san_francisco/businesses.csv \
| iconv -t UTF-8 -f ISO-8859-1 \
| cut -f 1 -d "," \
| sort \
| head -n 10
Adding uniq:
%%bash
tail -n +2 data/san_francisco/businesses.csv \
| iconv -t UTF-8 -f ISO-8859-1 \
| cut -f 1 -d "," \
| sort \
| uniq \
| wc
man uniq
There is one less unique business_id than there are lines in the file. Why is that?
In DS-100 we will largely focus on analysis within Python. However, as a data scientist you may want to learn more about using these Bash shell commands (common to all POSIX shells). Others worth learning are:
cd,ls,pwd,mkdir, andrmare file and folder traversal and manipulation commands.grepregular expression pattern matchingtrchange individual charactersteeused to split an output down two pathsawka string processing framework with it's own built in language for string transformations.zip,gzip,bzip2, andtarare compression and archiving commands
The Pandas DataFrame Library¶
In this class we will frequently operate on data stored in table form. A common data structure for tabular data in analysis tools is a DataFrame.
Conceptually DataFrames behave like tables (in a Spreadsheet) with rows corresponding to data records and columns corresponding to fields. DataFrames are also like Matrices in that we can index a row or column of data. However, unlike Matrices DataFrames can store data of different types in each column.
Many of you may already be familiar with DataFrames from Data8. In Data8 you used a DataFrame library called DataScience created here at Berkeley. The Berkeley DataScience library contains a simple DataFrame data structure with a few basic operations. In contrast many of the DataFrame APIs in use today (e.g., Pandas, Spark, and R) have fairly extensive APIs with numerous short-cuts and optimizations.
One of the most commonly used DataFrame APIs today is the Python Pandas Library. This library was heavily inspired by the DataFrame concepts in the R programming language.
In DS100 we will the Pandas DataFrame library for many of the assignments. We encourage students to read a little about Pandas here
Loading the DataFrame¶
One of the useful features of Pandas is its ability to load fairly messy "CSV" files:
import pandas as pd
# note the need to provide encoding information for this text file
bus = pd.read_csv("data/san_francisco/businesses.csv",
encoding='ISO-8859-1')
ins = pd.read_csv("data/san_francisco/inspections.csv")
vio = pd.read_csv("data/san_francisco/violations.csv")
If you look at the pd.read_csv command you will see that it has a very large number of arguments:
pd.read_csv?
To get a sense as to how the data was processed lets look at the records in each of the DataFrame
print('Records:', len(bus))
bus.head()
print('Records:', len(ins))
ins.head()
print('Records:', len(vio))
vio.head()
Now that we have successfully loaded the data let's start to examine what we have.
Questions about data organization?¶
- How is the data organized?
- What are the relationships between the tables (e.g., common attributes, keys)?
- Are there common data issues (e.g., missing values (NaN), date parsing)?
- How far does the data go back?
- others...
print("Bus(" + ", ".join(bus.columns) + ")")
print("Ins(" + ", ".join(ins.columns) + ")")
print("Vio(" + ", ".join(vio.columns) + ")")
The business_id field is shared across tables¶
The business_id field connects records across each of the DataFrames. Later in the class we will discuss this form of foreign key relationship between tables. For now, we will construct a modified version of these tables that are indexed by the business_id. This will both speedup future operations and also provide an implicit join key.
bus.set_index('business_id', inplace=True)
ins.set_index('business_id', inplace=True)
vio.set_index('business_id', inplace=True)
vio.head()
Date Parsing¶
Both the inspections and violations tables have dates.
vio['date'].head(10)
Notice the "type" of information stored in that column is an integer not a date. We need to clean that up a bit.
ins['date'] = pd.to_datetime(ins['date'], format="%Y%m%d")
vio['date'] = pd.to_datetime(vio['date'], format="%Y%m%d")
vio['date'].head(10)
How far back do the inspections and violation reports go:¶
print("Oldest inspection:", ins['date'].min())
print("Oldest violation:", vio['date'].min())
print("Most recent inspection:", ins['date'].max())
print("Most recent violation:", vio['date'].max())
Digging into the Inspection Data¶
ins.head()
It seems like there are possibly many routine inspections. Let's look at the distribution of inspection types. To do this we are going to select the type column and use the value_counts() function to count the occurrences:
ins['type'].value_counts()
Strange! There is only one complaint in the entire database.
Investigating the Complaint (Skip in Class)¶
Let's look a little bit further at this complaint:
ins[ins['type'] == 'complaint']
A score of 100?! That is odd. What was the restaurant?
ins[ins['type'] == 'complaint'].join(bus)
This probably deserves further investigation ...
vio.loc[87440]
vio.loc[87440].iloc[1,1]
And it still got a score of 100? This record is probably an anomaly but it does cause us to question the meaning of these scores.
How frequently are these restaurants inspected?¶
Group the inspections by the index (the business_id)
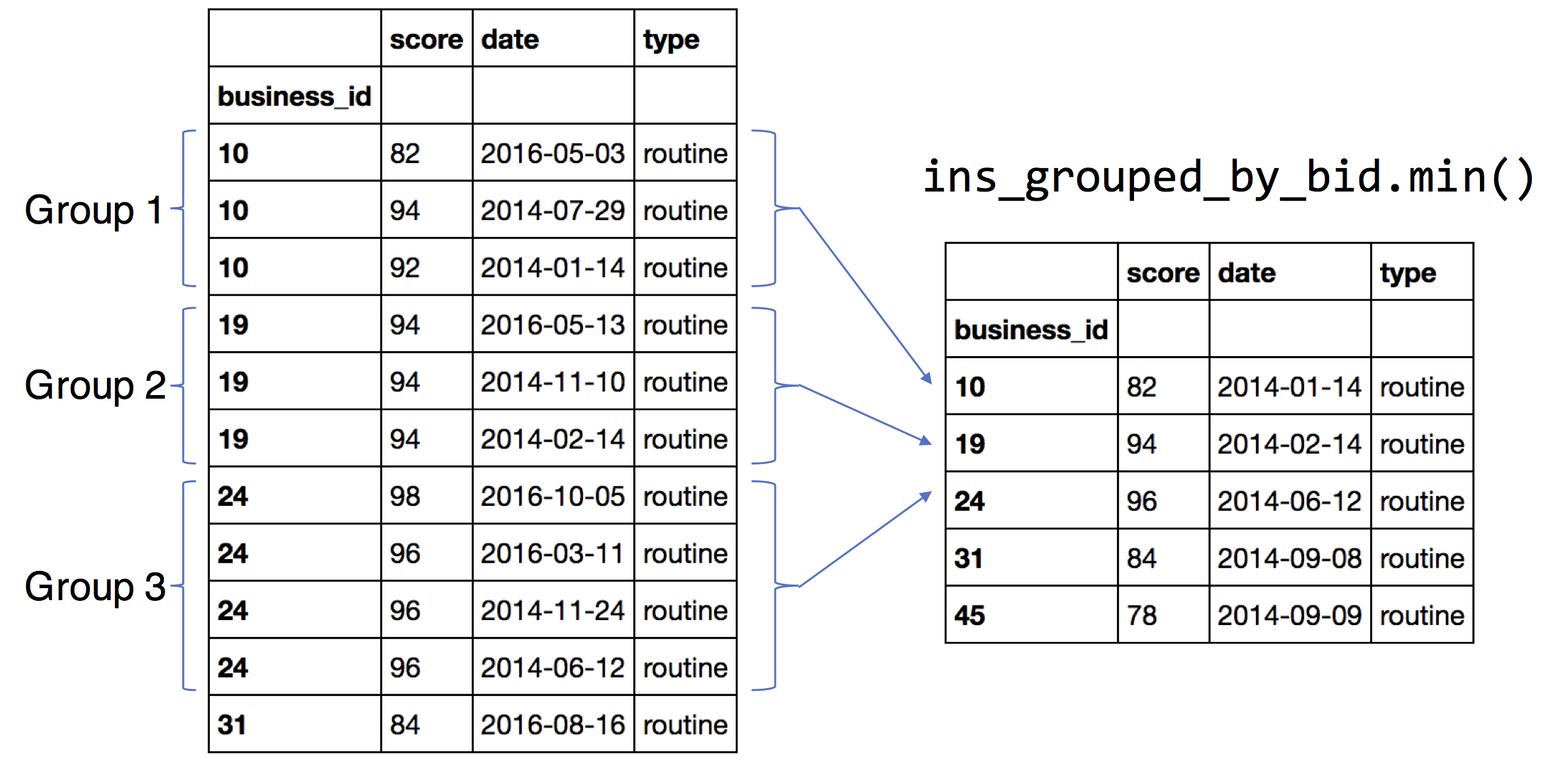
ins_grouped_by_bid = ins.groupby(ins.index)
bus['first_inspect'] = ins_grouped_by_bid['date'].min()
bus['last_inspect'] = ins_grouped_by_bid['date'].max()
bus['num_inspect'] = ins_grouped_by_bid['date'].count()
bus['days_bt_ins'] = (bus['last_inspect'] - bus['first_inspect']).dt.days / (bus['num_inspect'] - 1)
bus['years_bt_ins'] = bus['days_bt_ins'] / 365.0
bus[['years_bt_ins', 'days_bt_ins', 'first_inspect', 'last_inspect', 'num_inspect']].head(3)
%matplotlib inline
import seaborn as sns
import matplotlib.pyplot as plt
bus['years_bt_ins'].hist(bins=50)
What patterns do we see?
- Two modes at 1.0 and 2.0. Why?
Inspections are required annually. Why longer than 1 year?
Understanding Scores¶
Our original goal was to understand scores. Let's see what we can find. Since restaurants can have multiple inspections let's look at the most recent.
def most_recent(df):
return df.sort_values('date', ascending=False).iloc[0]
bus['latest_score'] = ins_grouped_by_bid.apply(most_recent)['score']
bus['mean_score'] = ins_grouped_by_bid['score'].mean()
Let's look at the spread of scores.
bus['latest_score'].hist(bins=10)
bus['latest_score'].hist(bins=200)
The scores seem to be pretty heavily skewed towards 100. Is that good news? There also appears to be a weird clustering around 90. Why?
Score inflation?¶
bus['latest_score'].describe()
What fraction of the restaurants are poor (below 70) by the cities standard?
threshold = 70
fraction = (bus['latest_score'] <= threshold).mean() # <- Why the mean?
print("Less than", str(round(fraction * 100, 2)) +
"% of the restaurants have a score of", threshold, "or less.")
Would you feel comfortable eating at a restaurant that got a score of 90?
What is the Geographic Distribution of Scores?¶
What data could we use to answer this question?
print("Bus(" + ", ".join(bus.columns) + ")\n")
print("Ins(" + ", ".join(ins.columns) + ")\n")
print("Vio(" + ", ".join(vio.columns) + ")")
- City?
- Postal Code
- Latitude & Longitude
Postal Codes (ZIP codes)¶
What is our coverage over postal codes?¶
What fraction of businesses don't have a postal code?
bus['postal_code'].isnull().mean()
Is that acceptable?
Let's look at how the businesses are spread over the postal codes.
zipcounts = bus['postal_code'].value_counts()
zipcounts.name = 'counts'
zipcounts.plot('bar')
What do we observe from above?¶
Observations?¶
- 94110 seems to have the most inspections.
- Messy Data!
CAis not a zipcode. - Some postal codes are in extended form.
Visualizing the counts of businesses in each postal code¶
Compute the number of occurrences for each postal code:
We will use the Folium mapping package. To install it make sure you are in the correct conda environment (e.g., ds100) and then use pip (not conda which is still a few versions behind):
pip install folium
import folium
folium.__version__
We need version 0.2.0 or greater.
import folium
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
# choropleth : Greek for choro=area + pleth=multitued
sf_map.choropleth(geo_path = "condensed_zipcodes.json",
data = zipcounts,
key_on = 'properties.zipcode',
threshold_scale = [100, 300, 400, 450, 500, 575],
fill_color='YlGn', fill_opacity=0.7, line_opacity=0.5)
sf_map
Get ZIP Code Labels and Areas¶
This requires the python area package. In your DS100 conda environment run:
pip
pip install area# Load my geo graphic data
import json
import numpy as np
import area
with open("condensed_zipcodes.json", "r") as f:
geojson = json.load(f)
def extract_info(x):
postal_code = x['properties']['zipcode']
coords = x['geometry']['coordinates'][0]
if len(coords) == 1:
coords = coords[0]
hectares = area.area(x['geometry']) / (10000.0)
return (postal_code, hectares, np.mean(coords, axis=0))
# Construct a dataframe with the number of hectares in each zipcode
postal_codes = pd.DataFrame(
[extract_info(g) for g in geojson['features']],
columns=['zipcode', 'hectares', 'coords']).set_index('zipcode')
postal_codes.head()
for zipcode, row in postal_codes.iterrows():
text = '<div style="font-size: 24pt">%s</div>' % zipcode
sf_map.add_child(folium.Marker(row['coords'][::-1], icon=folium.DivIcon(html=text)))
sf_map
How does this relate to the regions in SF?

Would we expect small zipcodes regions to have as many restaurants as big regions?
Normalizing by ZIP code by area¶
tmp = pd.DataFrame(zipcounts).join(postal_codes)
density = (tmp['counts'] / tmp['hectares']).dropna()
density.plot(kind='bar')
import folium
SF_COORDINATES = (37.77, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
#choropleth : Greek for choro=area + pleth=multitued
sf_map.choropleth(geo_path = "condensed_zipcodes.json",
data = density,
key_on = 'properties.zipcode',
fill_color='YlGn', fill_opacity=0.7, line_opacity=0.5)
sf_map
How does this relate to the regions in SF?

More Fine Grained Picture¶
We also have geo-coordinates attached to some of the restaurants. How many are missing?
How many of the entries are missing the latitude or longitude fields?
(bus['latitude'].isnull() | bus['longitude'].isnull()).mean()
We having missing values for 43% of the businesses. Is that a problem?
Nearly half are missing the latitude or longitude information! Let's look at the locations of those that do have a location.
bus_locations = bus[['name', 'latitude', 'longitude', 'latest_score', 'mean_score']].dropna()
print("There are", len(bus_locations), "businesses with locations.")
The following generates a HeatMap of locations where we have collected restaurant data with geo-location information.
import folium
import folium.plugins # The Folium Javascript Map Library
locs = bus_locations[['latitude', 'longitude']].as_matrix()
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
sf_map.add_children(folium.plugins.HeatMap(locs, radius = 10))
sf_map
Scores broken down by ZIP code¶
Focus on foodie ZIP codes¶
Lets define a foodie ZIP codes as one that has at least 200 restaurants.
# foodie_zips = zipcounts[zipcounts > 200].index
foodie_zips = density[density > 0.5].index
foodie_bus = bus[bus['postal_code'].isin(foodie_zips)]
foodie_bus.boxplot(by='postal_code', column='latest_score')
_ = plt.xticks(rotation='vertical')
Another kind of visualization (Skip)¶
sns.violinplot(x="postal_code", y="latest_score",
data=foodie_bus)
_ = plt.grid(b=True, which='major', color='w', linewidth=1.0)
_ = plt.xticks(rotation='vertical')
Why do the scores go above 100?
sns.violinplot(x="postal_code", y="latest_score",
data=foodie_bus,
cut= 0)
_ = plt.grid(b=True, which='major', color='w', linewidth=1.0)
_ = plt.xticks(rotation='vertical')
score_quantiles = bus[bus['postal_code'].isin(foodie_zips)][['postal_code', 'latest_score']].dropna()\
.groupby('postal_code')['latest_score'].quantile([0.1, 0.25, .5, 0.75, 0.9])
Visualizing the Scores by Zipcode¶
First we need to get the boundaries of the zip codes for our foody_zips
foodie_geojson = geojson.copy()
foodie_geojson['features'] = [f for f in geojson['features'] if f['properties']['zipcode'] in foodie_zips]
Median scores by zipcode¶
import folium
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
sf_map.choropleth(geo_str = json.dumps(foodie_geojson),
data = score_quantiles[:, 0.50],
key_on = 'properties.zipcode',
fill_color='BuPu', fill_opacity=0.7, line_opacity=0.5)
sf_map
Lower 10% Quantile¶
import folium
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
sf_map.choropleth(geo_str = json.dumps(foodie_geojson),
data = score_quantiles[:, 0.1],
key_on = 'properties.zipcode',
fill_color='BuPu', fill_opacity=0.7, line_opacity=0.5)
sf_map
ZIP Code with most spread (Interquartile Range)¶
import folium
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
sf_map.choropleth(geo_str = json.dumps(foodie_geojson),
data = score_quantiles[:, 0.75] - score_quantiles[:, 0.25],
key_on = 'properties.zipcode',
fill_color='BuPu', fill_opacity=0.7, line_opacity=0.5)
sf_map
Low Scoring Locations¶
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
cluster = folium.MarkerCluster()
for _, row in bus_locations[bus_locations['latest_score'] < 70.0].iterrows():
cluster.add_child(folium.Marker(location=[row['latitude'], row['longitude']],
popup=(row['name'] +": " + str(row['latest_score']))))
sf_map.add_child(cluster)
sf_map
sf_map
What kinds of violations are correlated with low scores?¶
vio.head()
vio['description'].value_counts()
Dirty data!¶
It looks like the description field has data information! How many descriptions are there?
len(vio['description'].value_counts())
We can use regular expressions to extract dates and remove the:
[ date violation corrected: ##/#/#### ]
string
vio['resolved'] = pd.to_datetime(vio['description'].str.extract("(\d+\/\d+\/\d+)", expand=False))
vio['desc'] = vio['description'].str.replace("\[.*\]","").str.strip()
Now let's look at all the value counts
len(vio['desc'].value_counts())
print(vio['desc'].value_counts().to_string())
Restaurants with many violations might be a bad sign:
bus['num_violations'] = vio.index.value_counts()
What are the worst offenders?
bus.sort_values('num_violations', ascending=False)[['name', 'num_violations', 'postal_code']].head(10)
That might not be fair if the restaurant is old. Let's look at at the violations on the latest inspection.
vio_bus = vio.join(bus['last_inspect'])
recent_vio = vio_bus[vio_bus['date'] >= vio_bus['last_inspect']]
bus['num_recent_violations'] = recent_vio.index.value_counts()
bus.sort_values('num_recent_violations', ascending=False)[['name', 'num_violations', 'num_recent_violations']].head(10)
What about unresolved recent violations?
bus['num_unresolved_violations'] = recent_vio[recent_vio['resolved'].isnull()].index.value_counts()
bus.sort_values('num_unresolved_violations', ascending=False)[[
'name', 'num_violations', 'num_recent_violations','num_unresolved_violations']].head(10)
Lets Look at these violations on a map¶
75th Quantile Number of Recent Violations¶
score_quantiles2 = bus[['postal_code', 'num_recent_violations']].dropna()\
.groupby('postal_code')['num_recent_violations'].quantile([0.25, .5, 0.75])
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
sf_map.choropleth(geo_str = json.dumps(foodie_geojson),
data = score_quantiles2[:, 0.75],
key_on = 'properties.zipcode',
fill_color='BuPu', fill_opacity=0.7, line_opacity=0.5)
sf_map
SF_COORDINATES = (37.76, -122.45)
sf_map = folium.Map(location=SF_COORDINATES, zoom_start=12)
bus_loc = bus[~bus['latitude'].isnull()]
for bid, row in bus_loc[bus_loc['num_recent_violations'] > 7].iterrows():
msg = "[" + str(bid) + "] " + row['name'] +": " + str(row['latest_score']) + "--> " + \
" -- ".join(recent_vio.loc[bid]['desc'].values)
sf_map.add_child(folium.Marker(location=[row['latitude'], row['longitude']],
popup=msg))
sf_map
What are the Violations of high-scoring restaurants?¶
Recall:
bus['latest_score'].dropna().describe()
Let's look at restaurants that scored above 96
tmp = recent_vio.join(bus['latest_score'])
tmp[tmp['latest_score'] >= 96.].groupby('desc')['desc'].count().sort_values(ascending=False)
Violations of Low Scoring Restaurants¶
tmp = recent_vio.join(bus['latest_score'])
tmp[tmp['latest_score'] <= 70.].groupby('desc')['desc'].count().sort_values(ascending=False)
If you have the same violations do you have the same score?¶
features = pd.get_dummies(recent_vio['desc']).groupby(recent_vio.index).sum()
# training_data = bus[['latest_score', 'num_recent_violations']]\
# .join(features)
training_data = bus[['latest_score']].join(features)
training_data = training_data.dropna()
grouped_by_vio = training_data.groupby(list(features.columns))
res = pd.DataFrame({
'count': grouped_by_vio['latest_score'].count().values,
'mean': grouped_by_vio['latest_score'].mean().values,
'var': grouped_by_vio['latest_score'].var().values
})
res[res['count'] > 25]
Building a Predictive Model¶
We could try to build a regressive model to identify how violations might contribute to the final score.
X = training_data.drop('latest_score', axis=1).as_matrix()
Y = training_data['latest_score'].as_matrix()
How much data do we have?
print(X.shape)
import sklearn
import sklearn.model_selection
(Xtr, Xte, Ytr, Yte) = sklearn.model_selection.train_test_split(X,Y, test_size=.1)
import sklearn.linear_model
#model = sklearn.linear_model.LinearRegression()
model = sklearn.linear_model.LassoCV(eps=0.000001, n_alphas=1000, max_iter=50000, fit_intercept=True)
model.fit(Xtr, Ytr)
print("The R^2 value:", model.score(Xte, Yte))
Listing the coefficient of the model¶
The most negative coefficient determine the the violations that most contributed to a reduction in score.
feature_weights = pd.DataFrame({
'features': training_data.drop('latest_score', axis=1).columns,
'weights': model.coef_})
print(feature_weights.sort_values('weights'))
Done! (Not Really)¶
There are a lot of questions we left unanswered. The data for other cities is available and we didn't even begin to answer the questions about how food safety is changing.
